FlyEye Silhouette turns microscopy data into cell-specific measurements by finding contours that enclose each nucleus. The process involves several steps, each of which are briefly explained in the sections below. For a more detailed explanation, please refer to the methods section our manuscripts.
Channel selection
The first step is to use the Channel drop-down menu to specify which fluorescence channel marks cell nuclei. This would be the color channel that provides maximal contrast between nuclei and the background. In the example below, we’ve selected the ‘Blue’ channel that reflects the DAPI stain.
Image preprocessing
The specified image channel is then pre-processed to eliminate artifacts and reduce the effects of noise. Pre-processing entails three sequential operations, each of which may be tuned using parameters provided by Silhouette:
- Gaussian blur is used to smooth out sharp pixel-wise fluctuations in the image.
- Contrast-limited adaptive histogram equalization (CLAHE) is used to equalize the intensity of the nuclear stain throughout the image field.
- Mean shift filtration is then used to further smooth the image.
Contour detection
The pre-preocesssed images are then used to define an initial set of contours that enclose the entire image foreground. To identify the foreground, the image is binarized by thresholding all pixels by the image-wide mean intensity level. A morphological opening is performed to help induce separation between distinct objects in the foreground. Contours are then constructed along the boundaries that separate foreground and background regions. Pixels intensities are averaged within each contour in order to exclude any contours that fall below a user-defined threshold.
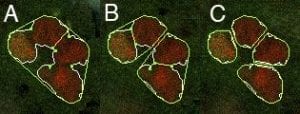
Contour splitting
Many contours may already enclose individual nuclei at this stage. However, in other cases a single contour may contain several nuclei. Silhouette uses a morphology-based approach to recursively split these composite contours into two or more subcomponents until each contour encloses a single nucleus.
At each level of recursion, a convex hull is constructed about each contour. A proposed cut-line is then drawn between the two contour vertices farthest from the convex hull. If the proposed cut meets a series of user-defined criteria, the contour is split into two adjacent contours. Recursion is repeated until either no more suitable cuts remain or a maximum recursion depth is reached.
Fluorescence quantification
Fluorescence intensities are then quantified for all image channels by evaluating the mean pixel intensity within each contour. The standard deviation is also computed for each fluorescence channel, along with the contour area and area centroid. All of these data are automatically saved within the Silhouette file container.